In a latest research revealed within the journal PNAS, researchers investigated how large-scale livestock rearing might outcome within the emergence and transmission of novel, doubtlessly zoonotic pathogens. They mixed a number of traces of proof from molecular relationship, comparative genomic analyses, and phylogeography to look at how the rearing of pigs allowed for novel Streptococcus suis lineages, a few of that are able to zoonotic spillover. Their findings reveal how pathogens, together with S. suis, adapt to take advantage of substantial adjustments of their host inhabitants sizes and the way this, in flip, can not directly contribute to the emergence of novel, doubtlessly pandemic-scale, and zoonotic, extremely pathogenic strains of hitherto benign microbes.
 Examine: The emergence and diversification of a zoonotic pathogen from inside the microbiota of intensively farmed pigs. Picture Credit score: Dusan Petkovic / Shutterstock
Examine: The emergence and diversification of a zoonotic pathogen from inside the microbiota of intensively farmed pigs. Picture Credit score: Dusan Petkovic / Shutterstock
Livestock rearing and its results on pathogens
Over the previous few centuries, the human inhabitants explosion and its related want for livestock as meals and labor to assist within the agricultural business has resulted within the international upscaling of livestock rearing. The agricultural suggestions loop of intensive farming programs allows bigger livestock populations, which in flip calls for elevated crop manufacturing. This has resulted in livestock populations now exceeding the mixed populations of people and wild animals.
These practices, together with the long-distance transport of reared animals, have contributed to low-genetic variety and high-density livestock. This presents a really perfect recipe for outbreaks of pathogens able to wiping out hundreds of thousands of livestock with out the genetic capability for resistance, which, when transported, can infect not solely different livestock populations but in addition wild populations of the identical or comparable species.
Alarmingly, this cocktail of occasions is hypothesized to advertise the emergence of novel zoonotic pathogens, arising from pathogens leaping to new hosts and mutations in beforehand benign microbiota beforehand related to reared animals.
“This path to pathogen emergence could also be notably essential in intensive farming programs, the place massive inhabitants measurement and excessive inhabitants density might choose for traits related to pathogenicity, whereas biosecurity reduces the chance of novel pathogens coming into the inhabitants.”
Streptococcus suis is a ubiquitous microbiota part of the higher respiratory tract of pigs. Beforehand benign, intensive rearing of pigs within the 19th and 20th centuries, the microbe has been noticed to adapt to a extra pathogenic way of life. In 1954, the micro organism was implicated in widespread illness in pigs, and at present presents one of the vital widespread illnesses in piglets. Alarmingly, collected mutations have allowed the micro organism to zoonotically spill over to people, related to meningitis, arthritis, endocarditis, and septicemia, with sudden demise in each human and porcine hosts.
Following the primary human S. suis-related mortality in 1968, the micro organism has since led to massive outbreaks in China and presents one of many main causes of grownup septicemia and meningitis throughout Southeast Asia.
“Difficulties in figuring out the determinants of pathogenicity in S. suis have been attributed to its advanced pathogenesis and excessive degree of genetic variety. Few research have thought of virulence components in strains apart from ST 1, which is answerable for most circumstances of S. suis illness in each pigs and people worldwide.”
In regards to the research
Within the current research, researchers investigated the associations between intensive pig rearing, the emergence of novel S. suis lineages, and their potential for zoonotic spillover. They carried out a inhabitants genomic evaluation of over 3,000 bacterial samples derived from tonsil and nasal swabs from pigs and wild boar. They moreover collected contaminated blood from people and pigs affected by S. suis illness throughout North America, Europe, Asia, and Australia. They aimed to elucidate the emergence, geographic unfold, and diploma of diversification of pathogenic lineages of the micro organism.
The research dataset comprised 3,070 genomic isolates of S. suis samples derived from beforehand current revealed knowledge and picked up and sequenced as part of this challenge. This included 29 revealed reference genomes and assortment isolates from 15 international locations unfold throughout the 5 continents above. Sampling was carried out between 2014 and 2018. Isolates have been processed via the Illunima complete genome HiSeq 25000 sequencing pipeline, which was then used to construct a genomic library of isolates. Uncooked sequences have been quality-checked, cleaned, and used to generate de novo assemblies for polymorphism evaluations.
The pipeline described by Athey et al. was used for serotyping and sequencing-typing analyses. Generated genomes have been subsequently annotated to determine homologous genes and analyze pathogenicity-associated genomic islands. The PopPunk software program was then used to determine divergent genomes and classify them into lineages. The six most typical lineages thus recognized have been examined for temporal indicators utilizing a regression of root-to-tip distances towards the 12 months of isolate sampling. Lastly, ancestral state reconstructions have been used to deduce the geographic unfold of recognized lineages.
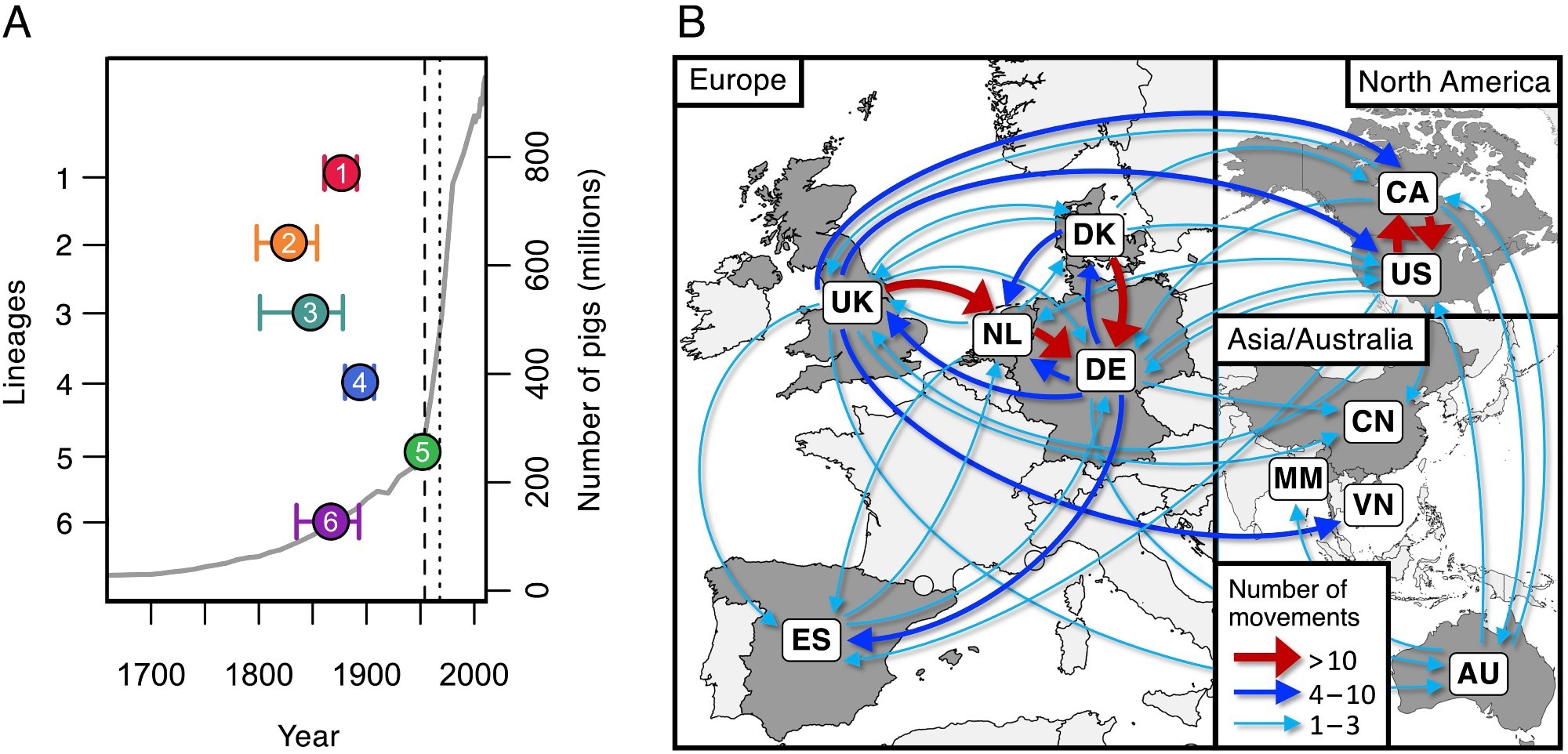 Dates of emergence and paths of between-country transmission for the six most typical pathogenic lineages. (A) Estimates of the dates of the latest widespread ancestors of the six most typical pathogenic lineages (coloured factors) towards an estimate of the worldwide variety of pigs (grey line). The vertical dashed line exhibits the date of the primary reported case of S. suis illness in pigs (1954), and the dotted line exhibits the primary reported human case (1968). (B) Map displaying inferred routes of transmission of those six pathogenic lineages between the international locations in our assortment. Arrows signify routes with at the least one inferred transmission occasion. Routes with greater than ten inferred transmission occasions are proven in purple, these with greater than three in blue, and people with one to 3 in turquoise.
Dates of emergence and paths of between-country transmission for the six most typical pathogenic lineages. (A) Estimates of the dates of the latest widespread ancestors of the six most typical pathogenic lineages (coloured factors) towards an estimate of the worldwide variety of pigs (grey line). The vertical dashed line exhibits the date of the primary reported case of S. suis illness in pigs (1954), and the dotted line exhibits the primary reported human case (1968). (B) Map displaying inferred routes of transmission of those six pathogenic lineages between the international locations in our assortment. Arrows signify routes with at the least one inferred transmission occasion. Routes with greater than ten inferred transmission occasions are proven in purple, these with greater than three in blue, and people with one to 3 in turquoise.
Examine findings
Examine findings revealed that over the previous 200 years, rearing porcine populations have elevated by over 200-fold, with the utmost enhance in the course of the latter half of the 20th century. These will increase have resulted within the admittedly gradual but regarding emergence of over 10 lineages with extremely pathogenic life histories. The excessive density of reared pigs, normally antibiotic-treated, has led to numerous, antibiotic-resistance lineages of S. suis presenting vital management challenges.
Analyses of samples from Spain reveal that pigs, each reared and wild boar, are hosts to S. suis strains which are extremely genetically numerous, suggesting that the affiliation between the microbe and its host has been long-standing. Genetic relationship analyses, nonetheless, revealed that all the six most typical pathogenic S. suis strains emerged in the course of the 19th and 20th centuries, corresponding with the unprecedented enhance in host rearing.
“The conclusion that these dates replicate an ecological shift towards pathogenicity in at the least a few of these lineages is supported by proof that they coincided with the acquisition of a pathogenicity-associated genomic island (Island 3). It’s additional supported by patterns of genome discount in every of the pathogenic lineages. In comparisons throughout bacterial species, it has been proven that bacterial pathogenicity is broadly related to smaller genomes and fewer genes.”
Analyses of the metabolic capacities of pathogenic lineages revealed that at the least two recognized genomic islands had considerably upped their capabilities for within-host progress, with all six islands depicting elevated metabolic actions over their extra benign counterparts. In different micro organism research, each within-host progress and metabolic charge have been linked to elevated virulence, suggesting a pattern in S. suis adapting to a extra virulent life historical past with larger potential for zoonotic spillover.
“Pathogenic lineages could also be higher in a position to exploit specific areas of the tonsil than commensal lineages and vice versa, thereby decreasing within-host competitors. This might result in segregation of those populations and diminished gene circulation between them, which might in flip result in the genome discount in additional pathogenic lineages on account of fewer alternatives for gene acquisition from extra numerous commensal lineages.”
Lastly, analyses revealed that present S. suis strains depict a excessive charge of unfold, able to quickly infecting a whole farm of pigs with the addition of 1 or a number of contaminated people. The massive-scale transport of livestock thereby presents a further downside: earlier endemic virulent strains being transmitted throughout nations and even continents, able to infecting novel host populations with little to no innate resistance towards them.
“Our outcomes present a framework for understanding the genomic variety in S. suis and its affiliation with pathogenicity. That is more likely to be of widespread use in S. suis analysis and in informing methods for controlling the burden of this illness on pig farming and human well being. As our assortment spans solely a small proportion of the international locations that farm pigs globally, additional sampling from a broader vary international locations and extra intensive sampling inside international locations, notably these with massive and rising pig populations, is required to research the existence of extra pathogenic lineages which are geographically restricted or have not too long ago emerged.”

