Kidney organoids present a promising platform in vitro to mannequin the mechanisms of kidney illness, nonetheless, they’re restricted by an present lack of information of their inherent useful protein expression. In a brand new report in Nature Communications, Martiz Lassé and a workforce of scientists in drugs and kidney well being, in Denmark, Germany, and the U.S., outlined the organoid proteome and their transcriptome trajectories throughout tradition, and upon publicity to the tumor necrosis issue alpha (TNFα); a cytokine stressor.
Whereas older organoids more and more deposited an extracellular matrix and expressed decreased glomerular proteins, the workforce targeted on extremely aggregated expressions of TNFα related protein expression in human tubular and organoid kidney cell populations to emphasise the scope of the fashions to advance the event of related biomarkers. The outcomes present essential proof of the useful relevance of the kidney organoid mannequin to recreate human kidney illness within the lab.
The organoid mannequin system
Organoids type an integral in vitro mannequin system to discover illness growth. Kidney organoids have modeled kidney most cancers, glomerular ailments, and polycystic kidney illness. These platforms additionally supply a promising screening device for therapeutics, to mannequin viral an infection and organ cryopreservation together with organ-on-a-chip microfluidics.
The idea can enhance the understanding of genetic kidney ailments, interrogate kidney growth and molecular mechanisms underlying pathologies. Kidney organoids could be generated from human pluripotent stem cells to recapitulate the weather of pathology which can be missing in two-dimensional tissue tradition fashions and in animal fashions.
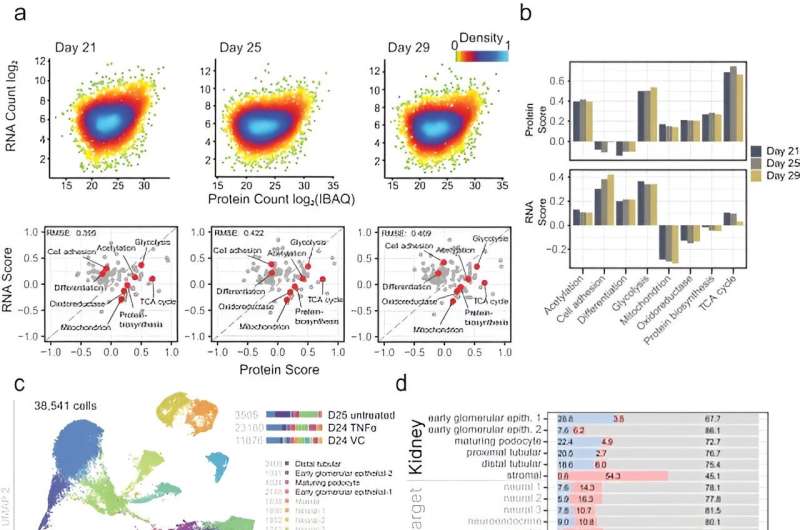
Through the experiments, Lassé and colleagues at first described the evolution of the organoid proteome and transcriptome as a perform of cell tradition period, to determine organoid cell sorts with altering protein expression. The workforce in contrast the expression of podocyte-specific proteins in organoids with the expression of native glomeruli and cultured podocyte cells.
The workforce analyzed the response of organoids to TNFα to supply disease-relevant biomarkers for precision diagnostics. Kidney organoid characterization allowed the platform’s growth for drug discovery, whereas assessing mobile biology underlying kidney illness to find potential therapies. The outcomes present a wealthy useful resource to the analysis group to advertise kidney illness modeling, biomarker discovery, and therapeutic screening.
The period of cell tradition and the altering kidney organoid protein expression ranges
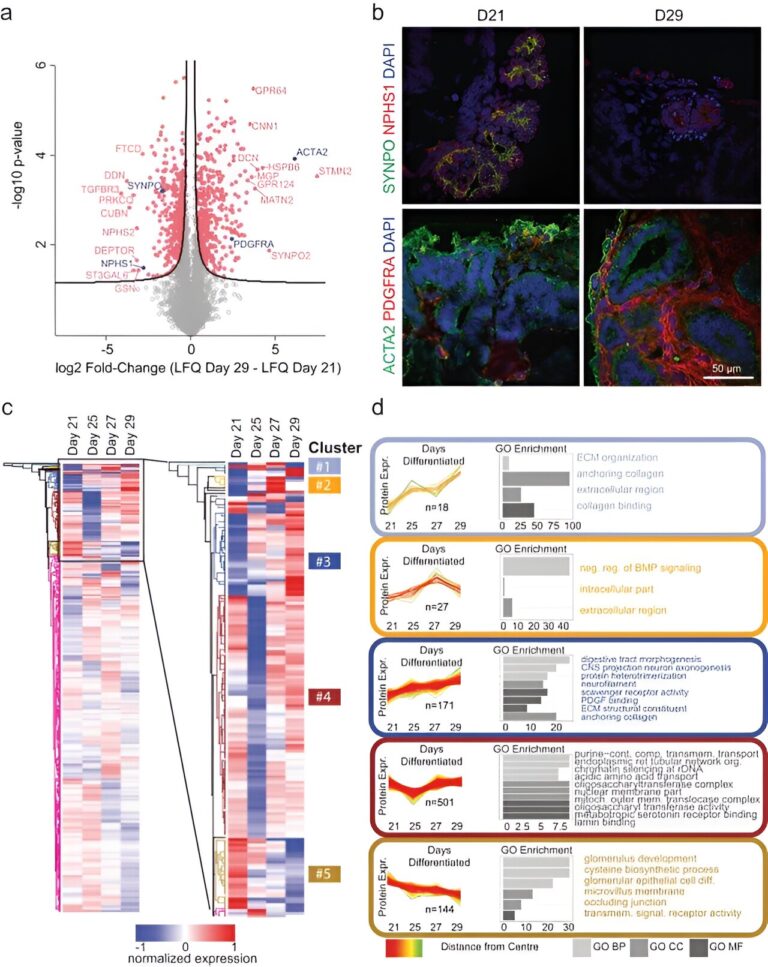
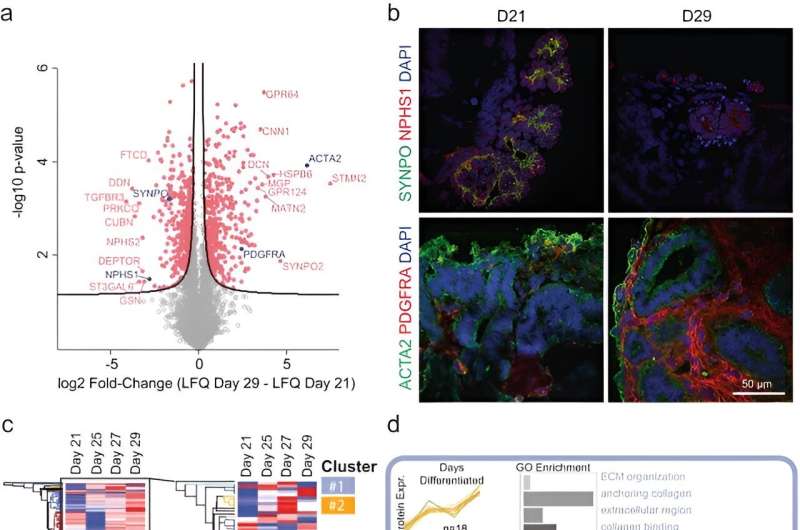
After three weeks of kidney organoid tradition, Lassé and colleagues carried out proteomic and transcriptional profiling. The scientists recognized and quantified 5,403 proteins utilizing principal part evaluation, the outcomes highlighted key podocyte markers/indicators of glomerular differentiation equivalent to nephrin and synaptopodin to lower with time, to spotlight a lack of podocytes or cell populations with time.
They noticed the elevated expression of structural proteins equivalent to easy muscle actin and key regulatory differentiation proteins platelet-derived progress issue receptor alpha, to spotlight an elevated manufacturing of the extracellular matrix. The scientists validated these observations with immunofluorescence. They equally famous larger expression of collagen kind I alpha chain and fibronectin kind 1 to substantiate the patterns of elevated extracellular matrix protein manufacturing with elevated period of cell tradition.
The experiments
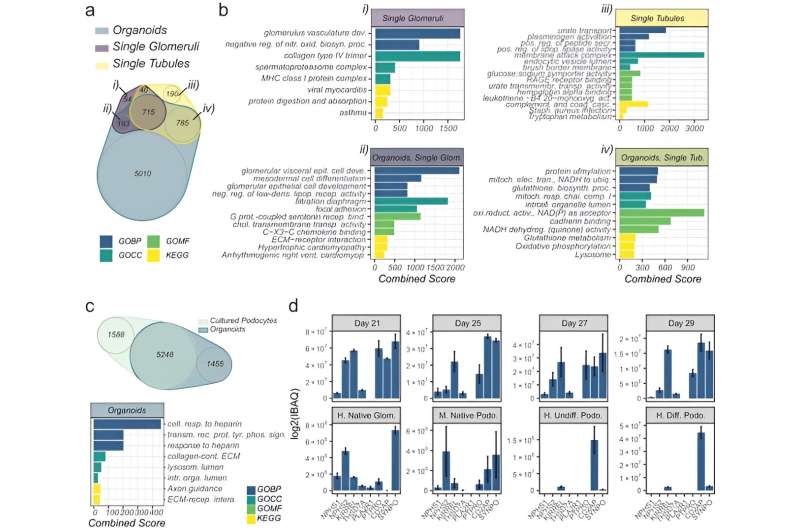
The scientists explored organoid proteome-transcriptome integration to uncover the mobile origins of proteins. The outcomes advised the most important proteome modifications to happen within the stromal cell inhabitants, whereas the early glomeruli epithelial cells and maturing podocytes had been much less represented with longer tradition period, in settlement with the decreased cell-specific protein expression evaluation too. The scientists in contrast the organoid proteome group with mature human kidney tissue, which confirmed similarities with native human kidneys to point how such kidney organoids faithfully represented most key proteins in native kidneys.
The position of TNFα in organoid tradition
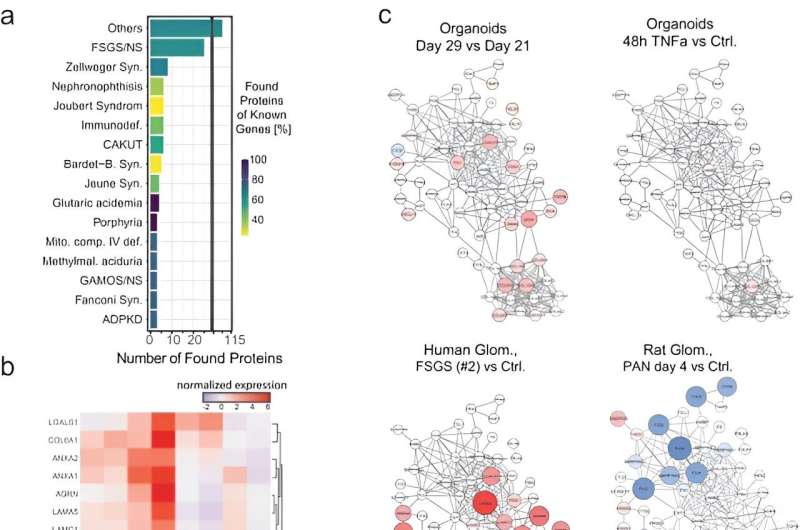
The workforce confirmed the response of organoids to TNFα by monitoring the expression of proinflammatory proteins. Notably, TNFα handled kidney organoids expressed key transcripts, together with chemokine ligand 2, chemoattractant protein 1, and the tissue inhibitor of metalloproteinases. Lassé and colleagues reproduced TNFα induced gene expression to indicate a posh organic response of handled organoids, in comparison with the human podocyte mannequin. The workforce demonstrated proteome alterations in TNFα induced organoids and revealed kidney cell-specific pathological mechanisms.
The TNF-dependent proteins additionally confirmed larger expression on a genetic degree in numerous diseased kidney tissue, when in comparison with dwelling donor tissue. Whereas on the mobile degree, the organoid TNF signature targeted on gene exercise, the comparative tissue TNF signature was extra advanced. Utilizing literature-based networks, the scientists explored the unified interaction of the networks to determine sub-groups of people in illness cohorts with numerous pathomechanisms benefiting from quite a lot of focused therapies.
Outlook
On this method, Martiz Lassé and colleagues confirmed how kidney organoids generated from human pluripotent stem cells have important scope to advance the understanding of kidney growth and illness. To efficiently implement this idea as a mannequin system, the scientists needed to perceive the advanced constructions and their relevance to human kidney illness. The experimental setup moreover supplied insights to the pathobiology of TNFα related kidney illness.
The examine outcomes help using organoids to mannequin advanced human kidney ailments. By integrating the omics knowledge derived from organoid illness fashions and different preclinical fashions, the workforce can drive higher kidney outcomes for sufferers. The organoids may also seize essential parts of advanced and heterogenous responses of the human kidney illness spectrum.
The omics map highlighted the capability to set off irritation and fibrosis in organoids, even within the absence of immune cells to open the door to mannequin extra advanced ailments with capability to personalize illness modeling in addition to therapeutic screening.
Extra data:
Moritz Lassé et al, An built-in organoid omics map extends modeling potential of kidney illness, Nature Communications (2023). DOI: 10.1038/s41467-023-39740-7
The Organoid Cell Atlas, Nature Biotechnology (2020). DOI: 10.1038/s41587-020-00762-x.
© 2023 Science X Community
Quotation:
Modeling the potential of kidney illness with an built-in organoid omics map (2023, September 4)
retrieved 4 September 2023
from https://medicalxpress.com/information/2023-09-potential-kidney-disease-organoid-omics.html
This doc is topic to copyright. Aside from any honest dealing for the aim of personal examine or analysis, no
half could also be reproduced with out the written permission. The content material is supplied for data functions solely.